To come to any great relation we require to connect the dots in the intelligent manner depending upon the previous knowledge inherited from the gene of our grandfathers.Since the future generation innovation is based on the knowledge base inherited from the DNA of their grandfathers free of cost,they are already at privileged state to start some great innovation.
Today again i will cover different topics in different domain and intelligently connect the dots to come to any conclusion.
So Start from Biological factors in our body and relate it to technology.
So our today's Biology topic is :
What is the genetic material?
The Hammerling Experiment: Cells Store Hereditary
Information in the Nucleus
Transplantation Experiments: Each Cell Contains a
Full Set of Genetic Instructions
The Griffith Experiment: Hereditary Information Can
Pass between Organisms
The Avery and Hershey-Chase Experiments: The
Active Principle Is DNA
What is the structure of DNA?
The Chemical Nature of Nucleic Acids. Nucleic acids
are polymers containing four nucleotides.
The Three-Dimensional Structure of DNA. The
DNA molecule is a double helix, with two strands
held together by base-pairing.
How does DNA replicate?
The Meselson–Stahl Experiment: DNA Replication Is
Semiconservative
The Replication Process. DNA is replicated by the
enzyme DNA polymerase III, working in concert
with many other proteins. DNA replicates by
assembling a complementary copy of each strand
semidiscontinuously.
Eukaryotic DNA Replication. Eukaryotic
chromosomes consist of many zones of replication.
What is a gene?
The One-Gene/One-Polypeptide Hypothesis. A gene
encodes all the information needed to express a
functional protein or RNA molecule.
How DNA Encodes Protein Structure. The
nucleotide sequence of a gene dictates the amino acid
sequence of the protein it encodes.
The realization that patterns of heredity can be explained by the segregation of chromosomes in meiosis
raised a question that occupied biologists for over 50 years: What is the exact nature of the connection between hereditary traits and chromosomes?
The Hammerling Experiment actually led to the conclusion that Cells Store Hereditary Information in the Nucleus.
Hereditary information in Acetabularia is stored in the foot of the cell, where the nucleus resides.
Transplantation Experiments: Each Cell Contains a Full Set of Genetic Instructions.
Hereditary information is stored in the nucleus of eukaryotic cells. Each nucleus in any eukaryotic cell contains a full set of genetic instructions.
What is the structure of DNA?
A German chemist, Friedrich Miescher, discovered DNA in 1869, only four years after Mendel’s work was published. Miescher extracted a white substance from the nuclei of human cells and fish sperm. The proportion of nitrogen and phosphorus in the substance was different from that in any other known constituent of cells, which convinced Miescher that he had discovered a new biological substance.
He called this substance “nuclein,” because it seemed to be specifically associated with the nucleus.
Levene’s Analysis: DNA Is a PolymerBecause Miescher’s nuclein was slightly acidic, it came to be called nucleic acid. For 50 years biologists did little research on the substance, because nothing was known of its function in cells. In the 1920s, the basic structure of nucleic acids was determined by the biochemist P. A. Levene, who found that DNA contains three main components (figure )
Nucleotide subunits of DNA and RNA. The nucleotide
subunits of DNA and RNA are composed of three elements: a
five-carbon sugar (deoxyribose in DNA and ribose in RNA), a
phosphate group, and a nitrogenous base (either a purine or a pyrimidine).
phosphate (PO4) groups; (2) five-carbon sugars; and (3) nitrogen-containing bases called purines (adenine, A, and guanine, G) and pyrimidines (thymine, T, and cytosine, C; RNA contains uracil, U, instead of T). From the roughly equal proportions of these components, Levene concluded correctly that DNA and RNA molecules are made of repeating units of the three components. Each unit, consisting of a sugar attached to a phosphate group and a base, is called a nucleotide. The identity of the base distinguishes one nucleotide from another. To identify the various chemical groups in DNA and RNA, it is customary to number the carbon atoms of the base and the sugar and then refer to any chemical group attached to a carbon atom by that number. In the sugar, four of the carbon atoms together with an oxygen atom form a five-membered ring. As illustrated in figure
Numbering the carbon atoms in a nucleotide. The
carbon atoms in the sugar of the nucleotide are numbered
1′ to 5′, proceeding clockwise from the oxygen atom. The
“prime” symbol (′) indicates that the carbon belongs to the
sugar rather than the base.
the carbon atoms are numbered 1′ to 5′, proceeding clockwise from the oxygen atom; the prime symbol (′) indicates that the number refers to a carbon in a sugar rather than a base. Under this numbering scheme, the phosphate group is attached to the 5′ carbon atom of the sugar, and the base is attached to the 1′ carbon atom. In addition, a free hydroxyl (—OH) group is attached to the 3′ carbon atom.
The 5′ phosphate and 3′ hydroxyl groups allow DNA and RNA to form long chains of nucleotides, because
these two groups can react chemically with each other. The reaction between the phosphate group of one nucleotide and the hydroxyl group of another is a dehydration synthesis, eliminating a water molecule and forming a covalent bond that links the two groups
The linkage is called a phosphodiester bond because the phosphate group is now linked to the two sugars by means of a pair of ester (P— O—C) bonds. The two-unit polymer resulting
from this reaction still has a free 5′ phosphate group at one end and a free 3′ hydroxyl group at the other, so it can link to other nucleotides. In this way, many thousands of nucleotides can join together
in long chains. Linear strands of DNA or RNA, no matter how long, will almost always have a free 5′ phosphate group at one end and a free 3′ hydroxyl group at the other. Therefore, every DNA and RNA molecule has an intrinsic directionality, and we can refer unambiguously to each end of the molecule. By convention, the sequence of bases is usually expressed in the 5′-to-3′ direction. Thus, the base sequence “GTCCAT” refers to the sequence, 5′ pGpTpCpCpApT—OH 3′ where the phosphates are indicated by “p.” Note that this is not the same molecule as that represented by the reverse sequence:
5′ pTpApCpCpTpG—OH 3′
Levene’s early studies indicated that all four types of DNA nucleotides were present
in roughly equal amounts. This result, which later proved to be erroneous, led to
the mistaken idea that DNA was a simple polymer in which the four nucleotides merely repeated (for instance, GCAT . . . GCAT . . . GCAT . . . GCAT . . .). If the sequence never varied, it was difficult to see how DNA might contain the hereditary information; this was why Avery’s conclusion that DNA is the transforming principle was not readily accepted at first. It seemed more plausible that DNA was simply a structural element of the chromosomes, with proteins playing the central genetic role.
Chargaff’s Analysis: DNA Is Not a Simple Repeating Polymer
When Levene’s chemical analysis of DNA was repeated using more sensitive techniques that became available after World War II, quite a different result was obtained. The four nucleotides were not present in equal proportions in DNA molecules after all. A careful study carried out by Erwin Chargaff showed that the nucleotide composition of DNA molecules varied in complex ways, depending on the source of the DNA . This strongly suggested that DNA was not a simple repeating polymer and might have the information-encoding properties genetic material must have. Despite DNA’s complexity, however, Chargaff observed an important underlying regularity in doublestranded DNA: the amount of adenine present in DNA always equals the amount of thymine, and the amount of guanine always equals the amount of cytosine. These findings are commonly referred to as Chargaff’s rules:
1. The proportion of A always equals that of T, and the proportion of G always equals that of C:
A = T, and G = C.
2. It follows that there is always an equal proportion of purines (A and G) and pyrimidines (C and T).
A single strand of DNA or RNA consists of a series of nucleotides joined together in a long chain. In all natural double-stranded DNA molecules, the proportion of A equals that of T, and the proportion of G equals that of C.
The Three-Dimensional Structure of DNA:Franklin: X-ray Diffraction Patterns of DNAThe significance of the regularities pointed out by Chargaff were not immediately
obvious, but they became clear when a British chemist, Rosalind Franklin carried
out an X-ray diffraction analysis of DNA. In X-ray diffraction, a molecule is bombarded with a beam of X
rays. When individual rays encounter atoms, their path is bent or diffracted, and the diffraction pattern is
recorded on photographic film. The patterns resemble the ripples created by tossing a rock into a smooth lake
When carefully analyzed,
they yield information about
the three-dimensional structure of amolecule. X-ray diffraction works best on substances that can be prepared as perfectly regular crystalline arrays. However, it was impossible to obtain true crystals of natural DNA at the time Franklin conducted her analysis, so she had to use DNA in the form of fibers. Franklin worked in the laboratory of British biochemist Maurice Wilkins, who was able to prepare more uniformly oriented DNA fibers than anyone had previously. Using these fibers, Franklin succeeded in obtaining crude diffraction information
on natural DNA. The diffraction patterns she obtained suggested that the DNA molecule had the shape of a helix, or corkscrew, with a diameter of about 2 nanometers and a complete helical turn every 3.4 nanometers
as shown below
Watson and Crick: A Model of
the Double Helix
Learning informally of Franklin’s results before they were published in 1953, James Watson and Francis
Crick, two young investigators at Cambridge University, quickly worked out a likely structure for the DNA molecule

which we now know was substantially correct. They analyzed the problem deductively, first building models of the nucleotides, and then trying to assemble the nucleotides into a molecule that matched what was known about the structure of DNA. They tried various possibilities before they finally hit on the idea that the molecule might be a simple double helix, with the bases of two strands pointed inward toward each other, forming base-pairs. In their model, basepairs always consist of purines, which are large, pointing toward pyrimidines, which are small, keeping the diameter of the molecule a constant 2 nanometers. Because hydrogen bonds can form between the bases in a base-pair, the double helix is stabilized as a duplex DNA molecule composed of two antiparallel strands, one chain running 3′ to 5′ and the other 5′ to 3′. The base-pairs are planar (flat) and stack 0.34 nm apart as a result of hydrophobic interactions, contributing to the overall stability of the molecule.
The Watson–Crick model explained why Chargaff had obtained the results he had: in a double helix,
adenine forms two hydrogen bonds with thymine, but it will not form hydrogen bonds properly with cytosine.
Similarly, guanine forms three hydrogen bonds with cytosine, but it will not form hydrogen bonds properly
with thymine. Consequently, adenine and thymine will always occur in the same proportions in any DNA molecule, as will guanine and cytosine, because of this base-pairing.
The Meselson–Stahl Experiment:DNA Replication Is Semiconservative
The Watson–Crick model immediately suggested that the basis for copying the genetic information is complementarity. One chain of the DNA molecule may have any conceivable base sequence, but this sequence completely determines the sequence of its partner in the duplex. For example, if the sequence of one chain is 5′- ATTGCAT-3′, the sequence of its partner must be 3′-TAACGTA-5′. Thus, each chain in the duplex is a complement of the other.The complementarity of the DNA duplex provides a ready means of accurately duplicating the molecule. If one were to “unzip” the molecule, one would need only to assemble the appropriate complementary nucleotides on the exposed single strands to form two daughter duplexes with the same sequence. This form of DNA replication is called semiconservative, because while the sequence of the original duplex is conserved after one round of replication, the duplex itself is not. Instead, each strand of the duplex becomes part of another duplex.Two other hypotheses of gene replication were also proposed. The conservative model stated that the parental double helix would remain intact and generate DNA copies consisting of entirely new molecules. The dispersive model predicted that parental DNA would become dispersed throughout the new copy so that each strand of all the daughter molecules would be a mixture of old and new DNA.The three hypotheses of DNA replication were evaluated
in 1958 by Matthew Meselson and Franklin Stahl of the California Institute of Technology. These two scientists grew bacteria in a medium containing the heavy isotope of nitrogen, 15N, which became incorporated into the bases of the bacterial DNA. After several generations, the
DNA of these bacteria was denser than that of bacteria grown in a medium containing the lighter isotope of nitrogen, 14N. Meselson and Stahl then transferred the bacteria from the 15N medium to the 14N medium and collected the DNA at various intervals. By dissolving the DNA they had collected in a heavy
salt called cesium chloride and then spinning the solution at very high speeds in an ultracentrifuge, Meselson and Stahl were able to separate DNA strands of different densities. The enormous centrifugal forces generated by the ultracentrifuge caused the cesium ions to migrate toward the bottom of the centrifuge tube, creating a gradient of cesium concentration, and thus of density. Each DNA strand floats or sinks in the gradient until it reaches the position where its density exactly matches the density of the cesium there. Because 15N strands are denser than 14N

The key result of the Meselson and Stahl experiment. These bands of DNA, photographed on the left and scanned on the right, are from the density-gradient centrifugation experiment of Meselson and Stahl. At 0 generation, all DNA is heavy; after one replication all DNA has a hybrid density; after two replications,
all DNA is hybrid or light. strands, they migrate farther down the tube to a denser
region of the cesium gradient. The DNA collected immediately after the transfer was all dense. However, after the bacteria completed their first round of DNA replication in the 14N medium, the density of their DNA had decreased to a value intermediate between 14N-DNA and 15N-DNA. After the second round of
replication, two density classes of DNA were observed, one intermediate and one equal to that of 14N-DNA.
The basis for the great accuracy of DNA replication is complementarity. A DNA molecule is a duplex,
containing two strands that are complementary mirror images of each other, so either one can be used as a
template to reconstruct the other.
The Replication Process
To be effective, DNA replication must be fast and accurate. The machinery responsible has been the subject of intensive study for 40 years, and we now know a great deal about it. The replication of DNA begins at one or more sites on the DNA molecule where there is a specific sequence of nucleotides called a replication origin
There the DNA replicating enzyme DNA polymerase III and other enzymes begin a complex process that catalyzes the addition of nucleotides to the growing complementary strands of DNA as shown below
How nucleotides are added in DNA replication. DNA polymerase III, along with other enzymes, catalyzes the addition of nucleotides to the growing complementary strand of DNA. When a nucleotide is added, two of its phosphates are lost as pyrophosphate.
DNA Polymerase III
The first DNA polymerase enzyme to be characterized, DNA polymerase I of the bacterium Escherichia coli, is a relatively small enzyme that plays a key supporting role in DNA replication. The true E. coli replicating enzyme, dubbed DNA polymerase III, is some 10 times larger and far more complex in structure. We know more about DNA polymerase III than any other organism’s DNA polymerase, and so will describe it in detail here. Other DNA polymerases are thought to be broadly similar. DNA polymerase III contains 10 different kinds of polypeptide chains, as illustrated in figure below
The DNA polymerase III complex. (a) The complex contains 10 kinds of protein chains. The protein is a dimer because both strands of the DNA duplex must be replicated simultaneously.
The catalytic (α) subunits, the proofreading (ε) subunits, and the “sliding clamp” (β2) subunits (yellow and blue) are labeled. (b) The “sliding clamp” units encircle the DNA template and (c) move it through the catalytic subunit like a rope drawn through a ring.

The enzyme is a dimer, with two similar multisubunit complexes. Each complex catalyzes the replication of one DNA strand. A variety of different proteins play key roles within each complex. The subunits include a single large catalytic α subunit that catalyzes 5′ to 3′ addition of nucleotides to a growing chain, a smaller ε subunit that proofreads 3′ to 5′ for mistakes, and a ring-shaped β2 dimer subunit that clamps the polymerase III complex around the DNA double helix. Polymerase III progressively threads the DNA through the enzyme complex, moving it at a rapid rate, some 1000 nucleotides per second (100 full turns of the
helix, 0.34 micrometers).
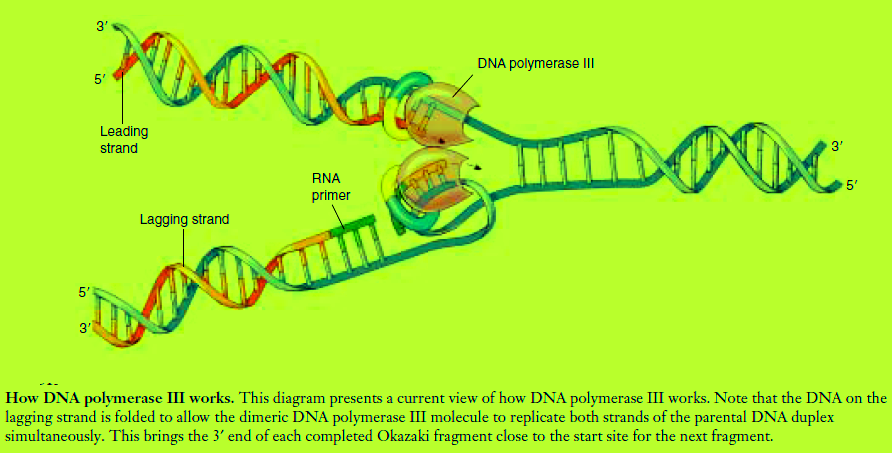
A DNA replication fork. Helicase enzymes separate the strands of the double helix, and single-strand binding proteins stabilize the single-stranded regions. Replication occurs by two mechanisms. (1) Continuous synthesis: After primase adds a short RNA primer, DNA polymerase III adds nucleotides to the 3′ end of the leading strand. DNA polymerase I then replaces the RNA primer with DNA
nucleotides. (2) Discontinuous synthesis: Primase adds a short RNA primer (green) ahead of the 5′ end of the lagging strand. DNA polymerase III then adds nucleotides to the primer until the gap is filled in. DNA polymerase I replaces the primer with DNA nucleotides, and DNA ligase attaches the short segment of nucleotides to the lagging strand.
The Two Strands of DNA Are Assembled in
Different Ways
The Need for a Primer
One of the features of DNA polymerase III is that it can add nucleotides only to a chain of nucleotides that is already paired with the parent strands. Hence, DNA polymerase cannot link the first nucleotides in a newly synthesized strand. Instead, another enzyme, an RNA polymerase
called primase, constructs an RNA primer, a sequence of about 10 RNA nucleotides complementary to the parent DNA template. DNA polymerase III recognizes the primer
and adds DNA nucleotides to it to construct the new DNA strands. The RNA nucleotides in the primers are then replaced by DNA nucleotides.
Another feature of DNA polymerase III is that it can add nucleotides only to the 3′ end of a DNA strand (the end with an —OH group attached to a 3′ carbon atom). This means that replication always proceeds in the 5′ → 3′ direction on a growing DNA strand. Because the two parent strands of a DNA molecule are antiparallel, the new strands are oriented in opposite directions along the parent templates at each replication fork (figure above) Therefore, the new strands must be elongated by different mechanisms! The leading strand, which elongates toward the replication fork, is built up simply by adding nucleotides continuously to its growing 3Œ end. In contrast, the lagging strand, which elongates away from the replication fork, is synthesized discontinuously as a series of short segments that are later connected. These segments, called Okazaki fragments, are about 100 to 200 nucleotides long in eukaryotes and 1000 to 2000 nucleotides long in prokaryotes. Each Okazaki fragment is synthesized by DNA polymerase III in the 5Œ ¨ 3Œ direction, beginning at the replication fork and moving away from it. When the polymerase reaches the 5Œ
end of the lagging strand, another enzyme, DNA ligase, attaches the fragment to the lagging strand. The DNA is further unwound, new RNA primers are constructed, and DNA polymerase III then jumps ahead 1000 to 2000 nucleotides (toward the replication fork) to begin constructing another Okazaki fragment. If one looks carefully at electron micrographs showing DNA replication in progress, one can sometimes see that one of the parent strands near the replication fork appears single-stranded over a distance of about 1000 nucleotides. Because the synthesis of the leading strand is continuous, while that of the
lagging strand is discontinuous, the overall replication of DNA is said to be semidiscontinuous.
The
Replication Process
The replication of the DNA double helix is a complex process that has taken decades of research to understand. It takes place in five interlocking steps:
1. Opening up the DNA double helix. The very stable DNA double helix must be opened up and its
strands separated from each other for semiconservative replication to occur. Stage one: Initiating replication. The binding of initiator proteins to the replication origin starts an intricate series of interactions that opens the helix. Stage two: Unwinding the duplex. After initiation,
“unwinding” enzymes called helicases bind to and move along one strand, shouldering aside the other
strand as they go. Stage three: Stabilizing the single strands. The unwound
portion of the DNA double helix is stabilized by single-strand binding protein, which binds to
the exposed single strands, protecting them from cleavage and preventing them from rewinding. Stage four: Relieving the torque generated by unwinding. For replication to proceed at 1000 nucleotides per second, the parental helix ahead of the replication fork must rotate 100 revolutions per second! To relieve the resulting twisting, called torque, enzymes known as topisomerases—or, more informally, gyrases—cleave a strand of the helix, allow it to swivel around the intact strand, and then reseal the broken strand. 2. Building a primer. New DNA cannot be synthesized on the exposed templates until a primer is constructed, as DNA polymerases require 3′ primers to initiate replication. The necessary primer is a short
stretch of RNA, added by a specialized RNA polymerase called primase in a multisubunit complex informally informally called a primosome. Why an RNA primer, rather than DNA? Starting chains on exposed templates introduces many errors; RNA marks this initial stretch as “temporary,” making this error-prone stretch easy to excise later.
3. Assembling complementary strands. Next, the dimeric DNA polymerase III then binds to the replication
fork. While the leading strand complexes with one half of the polymerase dimer, the lagging strand
is thought to loop around and complex with the other
half of the polymerase dimer (figure above). Moving in concert down the parental double helix, DNA
polymerase III catalyzes the formation of complementary sequences on each of the two single strands
at the same time.
4. Removing the primer. The enzyme DNA polymerase
I now removes the RNA primer and fills in the gap, as well as any gaps between Okazaki fragments.
5. Joining the Okazaki fragments. After any gaps between Okazaki fragments are filled in, the enzyme
DNA ligase joins the fragments to the lagging strand.
DNA replication involves many different proteins that open and unwind the DNA double helix, stabilize the single strands, synthesize RNA primers, assemble new complementary strands on each exposed parental strand—one of them discontinuously—remove the RNA primer, and join new discontinuous segments on the lagging strand.
Structure of Polynucleotide Chain:
Let us recapitulate the chemical structure of a polynucleotide chain (DNA or RNA). A nucleotide has three components – a nitrogenous base, a pentose sugar (ribose in case of RNA, and deoxyribose for DNA), and a phosphate group. There are two types of nitrogenous bases – Purines (Adenine and Guanine), and Pyrimidines (Cytosine, Uracil and Thymine). Cytosine is common for both DNA and RNA and Thymine is present in DNA. Uracil is present in RNA at the place of Thymine. A nitrogenous base is linked to the pentose sugar through a N-glycosidic linkage to form a nucleoside, such as adenosine or deoxyadenosine, guanosine or deoxyguanosine, cytidine or deoxycytidine and uridine or deoxythymidine. When a phosphate group is linked to 5'-OH of a nucleoside through phosphoester linkage, a corresponding nucleotide (or deoxynucleotide depending upon the type of sugar present) is formed. Two nucleotides are linked through 3'-5' phosphodiester linkage to form a dinucleotide. More nucleotides can be joined in such a manner to form a polynucleotide chain. A polymer thus formed has at one end a free phosphate moiety at

A Polynucleotide chain
5'-end of ribose sugar, which is referred to as 5’-end of polynucleotide chain. Similarly, at the other end of the polymer the ribose has a free 3'-OH group which is referred to as 3' -end of the polynucleotide chain.
The backbone in a polynucleotide chain is formed due to sugar and phosphates. The nitrogenous bases linked to sugar moiety project from the backbone In RNA, every nucleotide residue has an additional –OH group present at 2' -position in the ribose. Also, in RNA the uracil is found at the place of thymine (5-methyl uracil, another chemical name for thymine). DNA as an acidic substance present in nucleus was first identified by Friedrich Meischer in 1869. He named it as ‘Nuclein’. However, due to technical limitation in isolating such a long polymer intact, the elucidation of structure of DNA remained elusive for a very long period of time. It was only in 1953 that James Watson and Francis Crick, based on the X-ray diffraction data produced by Maurice Wilkins and Rosalind Franklin, proposed a very simple but famous Double Helix model for the structure of DNA. One of the hallmarks of their proposition was base pairing between
the two strands of polynucleotide chains. However, this proposition was also based on the observation of Erwin Chargaff that for a double stranded DNA, the ratios between Adenine and Thymine and Guanine and Cytosine are constant and equals one.
The base pairing confers a very unique property to the polynucleotide chains. They are said to be complementary to each other, and therefore if the sequence of bases in one strand is known then the sequence in other strand can be predicted. Also, if each strand from a DNA (let us call it as a parental DNA) acts as a template for synthesis of a new strand, the two double stranded DNA (let us call them as daughter DNA) thus, produced would be identical to the parental DNA molecule. Because of this, the genetic implications of the structure of DNA became very clear.
The salient features of the Double-helix structure of DNA are as follows:
(i) It is made of two polynucleotide chains, where the backbone is constituted by sugar-phosphate, and the bases project inside.
(ii) The two chains have anti-parallel polarity. It means, if one chain has the polarity 5'3', the other has 3'5' .
(iii) The bases in two strands are paired through hydrogen bond (H-bonds) forming base pairs (bp). Adenine forms two hydrogen bonds with Thymine from opposite strand and vice-versa. Similarly, Guanine is bonded with Cytosine with three H-bonds. As a result, always a purine comes opposite to a pyrimidine. This
generates approximately uniform distance between the two strands of the helix as shown in this new figure

(iv) The two chains are coiled in a right-handed fashion. The pitch of the helix is 3.4 nm (a nanometre is one billionth of a metre, that is 10-9 m) and there are roughly 10 bp in each turn. Consequently, the distance
between a bp in a helix is approximately equal to 0.34 nm.
(v) The plane of one base pair stacks over the other in double helix. This, in addition to H-bonds, confers stability of the helical structure the figure below:
Compare the structure of purines and pyrimidines. Can you find out why the distance between two polynucleotide chains in DNA remains almost constant? The proposition of a double helix structure for DNA and its simplicity in explaining the genetic implication became revolutionary. Very soon, Francis Crick
proposed the Central dogma in molecular biology, which states that the genetic information flows from DNARNAProtein.
In some viruses the flow of information is in reverse direction, that is, from RNA to DNA.
Packaging of DNA Helix
Taken the distance between two consecutive base pairs as 0.34 nm (0.34×10–9 m), if the length of DNA double helix in a typical mammalian cell is calculated (simply by multiplying the total number of bp with distance between two consecutive bp, that is, 6.6 × 109 bp × 0.34 × 10-9m/bp), it comes out to be approximately 2.2 metres. A length that is far greater than the dimension of a typical nucleus (approximately 10–6 m). How is such a long polymer packaged in a cell? If the length of E. coli DNA is 1.36 mm, can you
calculate the number of base pairs in E.coli? In prokaryotes, such as, E. coli, though they do
not have a defined nucleus, the DNA is not scattered throughout the cell. DNA (being negatively charged)
is held with some proteins (that have positive charges) in a region termed as ‘nucleoid’. The DNA
in nucleoid is organised in large loops held by proteins.
In eukaryotes, this organisation is much more complex. There is a set of positively charged, basic
proteins called histones. A protein acquires charge depending upon the abundance of amino acids
residues with charged side chains. Histones are rich in the basic amino acid residues lysines and
arginines. Both the amino acid residues carry positive charges in their side chains. Histones are
organised to form a unit of eight molecules called as histone octamer. The negatively charged DNA is wrapped around the positively charged histone octamer to form a structure called nucleosome. A typical nucleosome contains 200 bp of DNA helix. Nucleosomes constitute the repeating unit of a structure in nucleus called chromatin, thread-like stained (coloured) bodies seen in nucleus. The nucleosomes in chromatin are seen as ‘beads-on-string’ structure when viewed under electron microscope (EM)
Nucleosome show as following
EM picture - ‘Beads-on-String’
The beads-on-string structure in chromatin is packaged to form chromatin fibers that are further coiled and condensed at metaphase stage of cell division to form chromosomes. The packaging of chromatin at higher level requires additional set of proteins that collectively are referred to as Non-histone Chromosomal (NHC) proteins. In a typical nucleus, some region of chromatin are loosely packed (and stains light) and are referred to as euchromatin. The chromatin that is more densely packed and stains dark are called as Heterochromatin. Euchromatin is said to be transcriptionally active chromatin, whereas heterochromatin is inactive.
I will come to another topic in next page. watch my lectures on Physics,Chemistry and calculus.
https://www.youtube.com/channel/UCd2Gfi81vXUZiQlz6zQjVKQ
https://www.youtube.com/watch?v=daDUCxzfY00
https://www.youtube.com/watch?v=yNnIALKsJsw
https://www.youtube.com/watch?v=tkrR7bXv0s0
https://www.youtube.com/watch?v=w8E6rGVU_zk
https://www.youtube.com/watch?v=ixLYcmePnh8
https://www.youtube.com/watch?v=kGjIMyuwuwI
https://www.youtube.com/watch?v=Kz8WrjCcphs
https://www.youtube.com/watch?v=A16KsAvamco
Electronics And Communication check the link here:
http://electronicsandcommunicationadvancedma.blogspot.in/2015/03/3g-networksorthogonal-frequency.html
http://electronicsandcommunicationadvancedma.blogspot.in/2015/03/3g-networksorthogonal-frequency.html
http://electronicsandcommunicationadvancedma.blogspot.in/2015/03/3g-networksorthogonal-frequency.html